In medicine, the terms like remarkable and ground breaking seldom occur. However researchers from Cambridge University and the Institute of Computational Biology, Munich, Germany have done something beneficially remarkable by identifying 14 different diseases with common metabolic pathways and placing the information on an open access webserver.
As is well known in integrative medicine, multimorbidity requires a deep level of knowledge and a comprehensive approach to disease prevention. It is why integrative medicine is so critical to understanding how and why certain diseases lead to others. This new data tool, published in Nature Medicine, pairs metabolic pathways with multiple diseases. With one graph, doctors can easily see diseases that commonly occur together from the same metabolic pathway.
The recently published study and tool was developed by a team led by Dr. Claudia Langenberg, BIH Professor of Computational Medicine, and scientists from Munich and the United Kingdom. They have identified a number of metabolic processes that are associated with up to 14 diseases. The data came from more than 11,000 participants in the EPIC-Norfolk prospective cohort study, which records hundreds of measurements from blood samples, as well as clinical data on diseases over more than 20 years of follow-up.
“We wanted to know whether there are certain markers in the blood that indicate a risk, not only for one but for several diseases at the same time,” Langenberg explains.
To do this, the scientists first examined the concentration of hundreds of different molecules in the blood samples of a total of 11,000 study participants. They then examined how the concentration of individual metabolites was related to the onset of a total of 27 serious diseases in the participants. The metabolites included not only known metabolic products such as sugars, fats and vitamins, but also substances whose concentration depends on genetic or environmental factors. For example, the scientists were able to detect the degradation products of medications, coffee consumption or the presence of gut bacteria using a process known as “molecular profiling.”
By using blood samples from 20 years of electronic medical data and stored at minus 196 degrees Celsius since then. At that time, the people were mostly healthy. The diseases they developed afterwards were systematically recorded in detail for more than 20 years through electronic hospital data. “This allowed us to explore how the concentration of hundreds of molecules in the blood is linked to the development of one or multiple diseases,” Langenberg explains.
For example, the team found that the concentration of many metabolites in the blood that were associated with disease onset were explained by impaired liver and kidney function, obesity or chronic inflammation. But they also discovered that certain lifestyle factors or a reduced diversity of intestinal bacteria, also known as the gut microbiome, influence blood levels and can thus provide clues to the development of diseases over time.

It turned out that half of all detected molecules were associated with an increased or decreased risk of at least one disease – the majority with multiple, sometimes very different, diseases, pointing to metabolic pathways that increase the risk of multimorbidity.
Two-thirds of all Metabolites Associated with More than One Disease
“We found, for example, that an increased concentration of the sugar-like molecule N-acetylneuraminate increased the risk of no less than 14 diseases,” explains Maik Pietzner, a scientist working with Claudia Langenberg and lead author of the paper. “Gamma-glutamylglycine, on the other hand, is exclusively associated with the occurrence of diabetes. Other members of the same molecular groups simultaneously increase the risk of liver and heart disease.”
Langenberg adds: “Overall, we observed that two-thirds of the molecules are associated with the occurrence of more than one disease. This is in line with the fact that patients often develop a range of diseases in the course of their lives. If we succeed in influencing these key factors, this could make it possible to counter multiple diseases simultaneously.”
The following is an example of how the diseases are illustrated in the publically available tool, omicscience.org. The following shows how small molecule profiling identifies shared and distinct pathways to non-communicable disease multimorbidity.
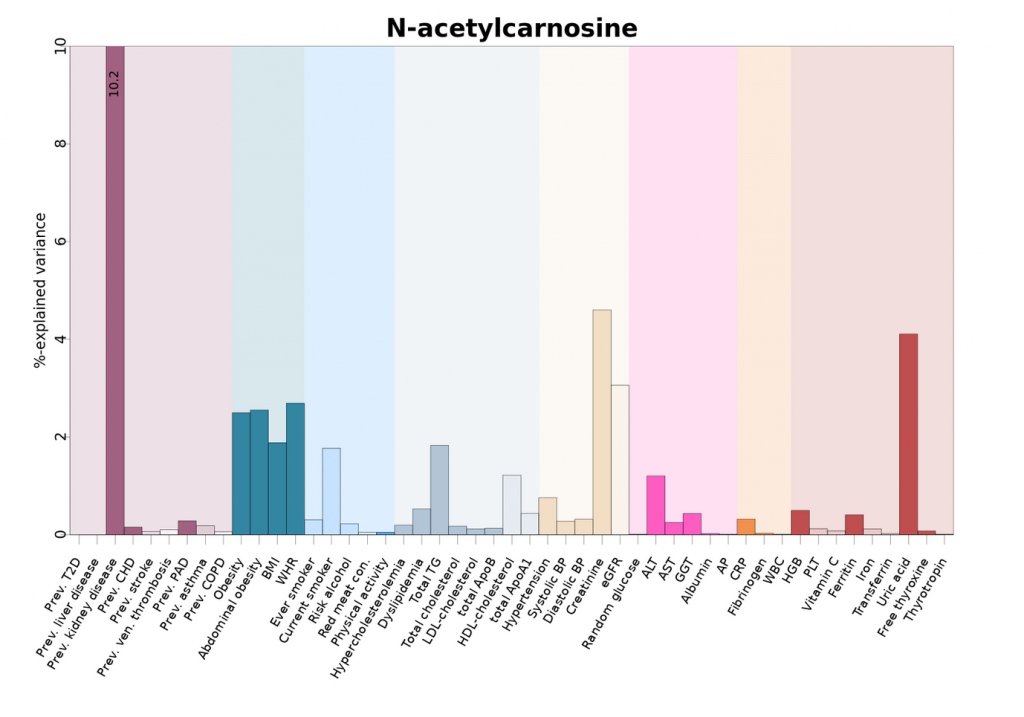
All results are publicly available
The scientists’ extensive analysis enables insights into the various factors influencing human metabolism that were previously not possible in this level of detail. To make this reference available to scientists around the world, the authors have developed a web application called omicscience.org / https://omicscience.org It makes all the results publicly available in graphical form so that they can be used in new studies. Langenberg adds: “The website enables scientists to determine key influencing factors for any molecule they are interested in or to uncover completely new connections between diseases. All of this was only possible due to our systematic, data-driven approach.”
NOTE: The researchers also integrated genomic and proteomic data from more than 10,000 individuals to characterize the genetic architecture of host proteins reported to interact with SARS-CoV-2 or to be related to virus entry, the host hyper-immune or procoagulant response, or severity of COVID-19. Explore the results here.
Conclusion/ Researchers investigate the association of 1,014 blood metabolites with 27 incident diseases, all-cause mortality and disease multimorbidity in more than 11,000 participants, and identified shared and distinct pathways to non-communicable diseases and multimorbidity. This provides a comprehensive catalogue of risk factor – metabolite – disease associations by integrating data from over 50 modifiable clinical and other risk factors. Explore the results here.