According to current studies, SARS-CoV-2 comprises at least five different variants. COVID-19 variants differ in how the immune system responds to the infection. Researchers from the German Center for Neurodegenerative Diseases (DZNE) and the University of Bonn, together with other experts from Germany, Greece and the Netherlands, published the findings in the journal “Genome Medicine.”
Infection with SARS-CoV-2 can manifest in different ways: Many of those affected do not even seem to notice the presence of the virus in their bodies. In other cases, the effects can include flu-like symptoms and neurological disorders to severe and even life-threatening pneumonia.
“The classification of COVID-19 into mild and severe courses falls short. The disease is much more diverse, and for each affected person, one certainly would want a therapy that is tailored to fit. What helps one person may be ineffective for another,” said Dr. Anna Aschenbrenner, a scientist of the LIMES Institute at the University of Bonn and the DZNE’s Systems Medicine division.
“In this respect, it is obvious to want to understand what underlies these differences. If we can pin them down to scientific criteria and categorize patients accordingly, this increases the chances of effective treatment. We therefore took a look at the immune system. Because many studies are indicating that its response to infection with SARS-CoV-2 plays a crucial role in the course of COVID-19,” said Aschenbrenner, who is a member of the Cluster of Excellence “ImmunoSensation” of the University of Bonn.
White Blood Cells ID COVID-19 Variants
In light of this, a team led by Anna Aschenbrenner, along with colleagues in Germany and abroad, analyzed the blood of people with and without COVID-19. The samples came from 95 people distributed among Bonn, Athens and Nijmegen.
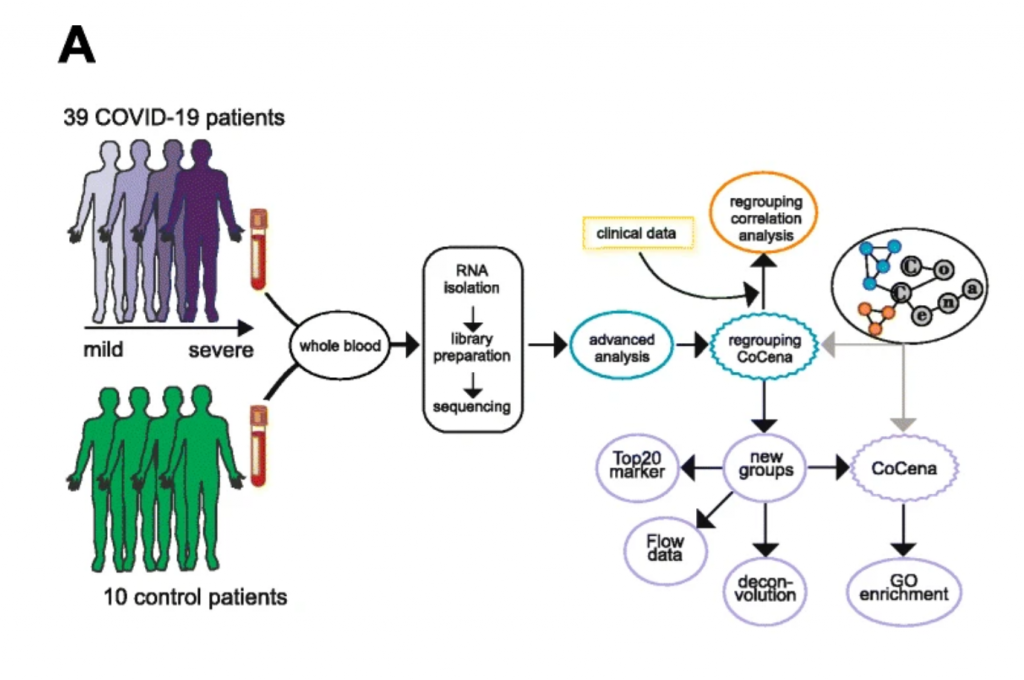
To investigate the host immune response of COVID-19 patients in a systematic approach, whole blood transcriptomes were analyzed from 39 patients and 10 control donors recruited at the same hospital by RNA-sequencing. Based on the molecular fingerprint generated in through the bioinformatics, the researchers identified which genes within the immune cells were switched on or off. The “expression patterns” provided information about the condition of cells and thus about their properties and functions, which can change depending on the situation.
Interestingly, the picture obtained in this way was largely determined by the family of “neutrophils”, which are the most abundant of the white blood cells and quite up front in the reaction chain of the immune response. These cells are thus mobilized very early to defend against infections. They act upon the formation of antibodies and, moreover, on other cells that contribute to immunity.
Study Highlights
- Neutrophil activation-associated signatures were prominently enriched in severe patient groups,
- This was corroborated in whole blood transcriptomes from an independent second cohort of 30 as well as in granulocyte samples from a third cohort of 16 COVID-19 patients (44 samples).
- Comparison of COVID-19 blood transcriptomes with those of a collection of over 3100 samples derived from 12 different viral infections, inflammatory diseases, and independent control samples revealed highly specific transcriptome signatures for COVID-19.
- Further, stratified transcriptomes predicted patient subgroup-specific drug candidates targeting the dysregulated systemic immune response of the host.
Five Manifestations that Differ From Healthy People
“First of all, it is important to note that the expression patterns of immune cells in people with COVID-19 differ fundamentally from those in healthy individuals. The gene activity we can detect in the blood is strongly altered. But there are also striking differences among patients. On this basis, we have identified five different groups. We refer to them as molecular phenotypes,” said Dr. Thomas Ulas, an expert in bioinformatics at the DZNE.
“Two of them represent severe disease courses. The others have more moderate symptoms,” said Ulas. The classification was based solely on transcriptome data. Only in retrospect, molecular phenotypes were matched to registered clinical courses.
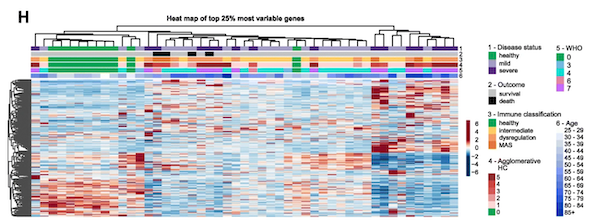
Interestingly, neither COVID-19 disease status, immune classification, nor our clinical parameter-based grouping of the COVID-19 patients aligned with overall transcriptional variability in the data (See Fig H below), indicating that hidden information in the blood transcriptome may guide further patient stratification.
How is COVID-19 Different from other Diseases?
The researchers used their findings to compare COVID-19 with other diseases and also with data from healthy individuals. For this purpose, they were able to draw on data from the “Rhineland Study” – a population study conducted by the DZNE in the Bonn area – as well as on data from scientific databases.
For the comparison, a large spectrum of diseases was considered: including viral infections such as influenza, infections with HIV and Zika, bacterial infections such as tuberculosis and bacterial sepsis, and inflammatory diseases such as rheumatoid arthritis.
“All five COVID-19 phenotypes are different from the other diseases we studied,” Ulas said, summing up the findings. “Apparently, COVID-19 has a unique biology that is reflected in the gene activity of immune cells in the blood. Insofar, expression analysis could be used to diagnose COVID-19. This would be an alternative or complement to current methods.”
Searching for Drugs for COVID-19 Variants
The scientists also searched for potential drugs against COVID-19. For this, they drew on the effects registered in databases of around 900 approved drugs on the expression patterns of cells. “We calculated which pharmaceuticals could counteract the altered gene activity profiles of the individual COVID-19 phenotypes,” said Aschenbrenner. On this basis, drug candidates for therapy were identified.
“Already in April of last year, we calculated a potential efficacy for example for dexamethasone and baricitinib in one of the patient groups with severe course that we identified. These types of analyses, it must be clearly stated, are not treatment recommendations. They do, however, very much provide starting points for therapy development, which then need to be tested in appropriate trials. In the case of dexamethasone and baricitinib, our predictions turned out to be correct. This is an indication of the strength of our approach of using blood transcriptomes to better characterize and classify patients,” said Aschenbrenner.
Conclusion/ “This study provides novel insights in the distinct molecular subgroups or phenotypes that are not simply explained by clinical parameters. We show that whole blood transcriptomes are extremely informative for COVID-19 since they capture granulocytes which are major drivers of disease severity.”
Click Here for Full Text Study




